In the recently published book,
America's Nuremberg: Human Experimentation
in Anabolic Steroid Research, Michael Scally, M.D. brings to light the ethical,
legal, and medical failures of the research community to recognize or investigate
the period after AAS cessation. Historically, the difference in the beliefs
held by the athletic and the physician/academic communities on AAS are contradictory
and irreconcilable. Illicit AAS users stated observations of the relationship
between AAS and muscle have proven to be true despite the rhetoric to the opposite.
This is true not only for the association between AAS and muscle but also for
the effects of AAS cessation upon muscle. The former took over sixty years for
the medical community to admit, while the medical community continues to fail
to recognize or investigate the latter. Rather than wait the sixty years it
took the medical community to awaken to its shortcomings this book will serve
the greater purpose of hastening their attention to the period after AAS cessation.
Now, in his forthcoming book Scally continues his first-rate investigative
and research abilities into producing an opus on steroid science. In the tradition
of supplying the well-educated and intelligent Meso readers with the best available
information available, Dr. Scally is providing Meso with summaries of the forthcoming
book. The article following is the first in a series that highlight selected
excerpts from this book. The androgen receptor is a member of the nuclear receptor
(NR) family. Research and investigations on the androgen receptor (AR) are creating
a revolution for ligands, steroidal and nonsteroidal, that produce anabolic
changes in muscle. To fully appreciate and understand these new drugs requires
knowledge for the nuclear receptor. The first of these articles introduces the
nuclear receptor. Following this will be an article on the androgen receptor,
introducing the all-important ligand-binding pocket (LBP). The third in the
series will document the molecular interactions of various ligands, steroidal
and nonsteroidal, for amino acid residues within the LBP.
THE NUCLEAR RECEPTOR
Multicellular organisms require specific intercellular communication to orchestrate
properly the complex body plan during embryogenesis and maintain the physiological
properties and functions during the entire lifespan. The binding of extracellular
signal molecules to either cell-surface receptors or intracellular receptors
differs. Most signal molecules are hydrophilic and are therefore unable to cross
the plasma membrane directly; instead, they bind to cell-surface receptors,
which in turn generate one or more signals inside the target cell. A number
of small hydrophobic signal molecules diffuse directly across the plasma membrane
of target cells and bind to intracellular receptor proteins. These signal molecules
include steroid hormones, thyroid hormones, retinoids, and vitamin D. Although
they differ greatly from one another in both chemical structure and function,
they all act by a similar mechanism.
Initiation of classical signal transduction cascades are by ligand binding
to membrane-anchored receptors, eventually changing the activity of specific
nuclear transcription factors. While growth factors, neurotransmitters, and
peptide hormones bind to membrane receptors thereby inducing the activity of
intracellular signaling pathways, other small hydrophobic signaling molecules
such as steroid hormones, certain vitamins, and metabolic intermediates enter
the target cells and bind to cognate members of a large family of nuclear receptors
(NR).[1] In contrast, members of
the nuclear receptor superfamily transduce their signals directly. The receptors
have evolved to combine the functions of signal responsiveness, DNA-binding,
and transcriptional activation into one protein composed of functionally separated
modules/domains.[2]
Nuclear receptors (NR) are members of a large superfamily of evolutionarily
related DNA-binding transcription factors that regulate programs involved in
a broad spectrum of physiological phenomena.[3]
Nuclear receptors regulate diverse functions, such as homeostasis, reproduction,
development, and metabolism. Nuclear receptors function as ligand-activated
transcription factors, and thus provide a direct link between signaling molecules
that control these processes and transcriptional responses.[4]
The NR is of major importance for intercellular signaling, converging intracellular
and extracellular signals on the regulation of genetic programs. They are transcription
factors that: (1) respond through association with a variety of hormonal and
metabolic signals, (2) integrate diverse signaling pathways as they correspond
themselves to targets of post-translational modifications, and (3) regulate
the activities of other major signaling cascades. The genetic programs that
they establish or modify affect virtually all aspects of the life, covering
such diverse aspects as embryogenesis, homeostasis, reproduction, cell growth,
or death.[5]
NR emerged in the earliest of metazoan evolution, long before the divergence
of vertebrates and invertebrates.[6]
The NR superfamily consists of a large group of transcription factors that are
essential for growth, differentiation, metabolism, reproduction, and morphogenesis.
More than fifty members of the NR superfamily all derive from a common ancestor.[7]
Complementary DNA (cDNA)[8] for many
intracellular receptors have been isolated and characterized, making it possible
to deduce the amino acid sequences of various nuclear receptors and related
members of the superfamily of nuclear receptors.[9]
The phylogenetic transparency of these receptors makes classification by sequence
relatively straightforward. Sequence alignment and phylogenetic tree construction
resulted in a classification of the human NR family into six evolutionary groups,
subfamilies, of unequal size.[10]
|
NUCLEAR RECEPTOR
SUPERFAMILY
|
|
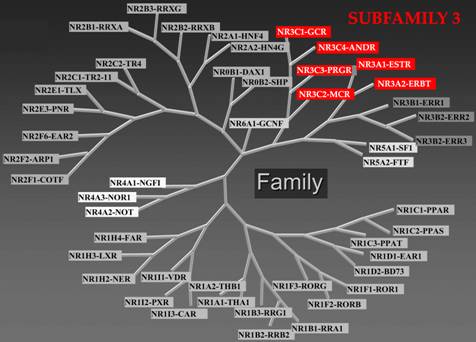
|
|
Nuclear receptor subfamily 3, Estrogen Receptor-like contains
Group A: Estrogen receptors include members 1: Estrogen receptor-α
(ERα; NR3A1, ESR1) and 2: Estrogen receptor-β (ERβ; NR3A2, ESR2);
Group B, the Estrogen related receptors include members 1: Estrogen
related receptor-α (ERRα; NR3B1, ESRRA), 2: Estrogen related receptor-β
(ERRβ; NR3B2, ESRRB), and 3: Estrogen related receptor-γ (ERRγ;
NR3B3, ESRRG); and Group C, the 3-Ketosteroid receptors include
members 1: Glucocorticoid receptor (GR; NR3C1) (Cortisol), 2: Mineralocorticoid
receptor (MR; NR3C2) (Aldosterone), 3: Progesterone receptor (PR;
NR3C3, PGR) (Sex hormones: Progesterone), and 4: Androgen receptor
(AR; NR3C4, AR) (Sex hormones: Testosterone). |
The steroid hormone receptors (SHR) subgroup of the nuclear receptor superfamily
is nuclear receptor subfamily 3. Subfamily 3A includes the estrogen receptor,
which comes in two forms, ERα [NR3A1] and ERβ [NR3A2]. Subfamily 3C includes
the cortisol binding glucocorticoid receptor (GR) [NR3C1], the aldosterone binding
mineralocorticoid receptor (MR) [NR3C2], the progesterone receptor (PR) [NR3C3],
and the dihydrotestosterone (DHT) binding androgen receptor (AR) [NR3C4]. In
addition, the SHR subgroup contains three orphan receptors closely related to
ER, subfamily 3B: the estrogen related receptors α (ERRα) [NR3B1], β (ERRβ)
[NR3B2], and γ (ERRγ) [NR3B3], for which a natural ligand remains to be identified.[11]
To date, no experimentally determined three-dimensional structure is available
for a complete receptor. It has so far not been possible to determine the tertiary
structure of intact full-length NR. Nevertheless, conformational
studies of NR parts yield valuable structural information. There
is a consistent structural and functional organization of the nuclear receptor
superfamily.
Cloning and sequencing of the genes encoding nuclear receptors permit comparison
of their amino acid sequences. Such studies revealed a remarkable conservation
in both the amino acid sequences and different functional regions of various
nuclear receptors. Nuclear receptors share a common structural organization.
NR is a modular protein with distinct functional and structural domains. The
members of the steroid hormone receptor family share a similar, modular architecture,
consisting of a number of independent functional domains.
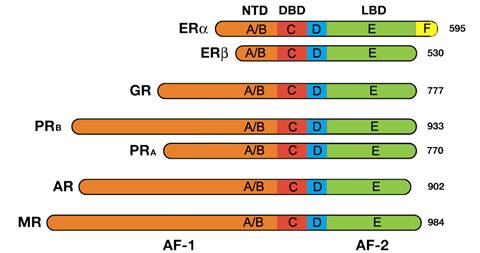
All steroid hormone receptors are composed of a variable N-terminal domain
(NTD, A/B); a highly conserved DNA Binding Domain (DBD, C); a flexible hinge
region (D); and a C-terminal Ligand Binding Domain (LBD, E). The estrogen receptor
α is unique in that it contains an additional C-terminal (F) domain with unknown
function.[12]
|

|
|
Nuclear receptors consist of six domains (A-F) based on regions
of conserved sequence and function.[13]
The evolutionarily conserved regions are C and E, and the divergent
regions A/B, D, and F regions.[14]
|
Nuclear receptors contain a variable N-terminal amino acid sequence, (NTD,
A/B), which contains an autonomous transcriptional activation function known
as AF-1. The AF-1 shows weak conservation across the nuclear receptor superfamily
and may mediate differential promoter regulation in vivo. The AF-1 sequence
functions as a ligand-independent transcriptional activator, but can also functionally
synergize with AF-2. The NTD is unique to each SHR and has variable sequence
and length. The highly conserved C region harbors the DNA-binding domain that
confers sequence-specific DNA recognition (DBD, C). Situated between the DBD
and the LBD is a linker region, domain D. This region functions as a flexible
hinge and contains the nuclear localization signal (NLS). The LBD (E) is responsible
for the binding of cognate ligand or hormone. This domain also contains a ligand-dependent
transcriptional activation function (AF-2) necessary for recruiting transcriptional
coactivators, which interact with chromatin remodeling proteins and the general
transcriptional activation machinery. Nuclear receptors may or may not contain
a final domain in the C-terminus, the F region, whose sequence is extremely
variable and whose structure and function are unknown.[15]
|
|
|
The domains starting from the N-terminus (left) to C-terminus
(right). NTD = N-terminal domain, DBD = DNA binding domain. LBD
= ligand binding domain. AF = activation function. The steroid hormone
receptor abbreviations are ER – estrogen receptor, GR – glucocorticoid
receptor, PR –progesterone receptor, AR – androgen receptor, and
MR – mineralocorticoid receptor. The numbers to the right are the
lengths in amino acid residues.
|
A/B REGION (N-TERMINAL DOMAIN, NTD)
Thus far, there is no elucidation of a crystal structure of an A/B domain.
The A/B region in the different NR is highly variable, revealing a very weak
evolutionary conservation. The N-terminal region is the least conserved
region among NR, both in size and sequence.[16]
All the nuclear receptors have a unique N-terminal region (NTD) of variable
length (100–500 amino acids) whose 3D structure is unknown.
NR contains two transactivation functions. One maps to the structurally flexible
N-terminal domain (NTD) and is termed AF-1. The poorly defined N-terminal A/B
region contains a transcriptional activation function, referred to as activation
function 1 (AF-1) that can operate autonomously. AF-1 can act in a ligand-independent
manner when placed outside of the receptor. Besides the one constitutionally
active transactivation region (AF-1), NTD includes several autonomous transactivation
domains (AD). The activation domains (AD) contain transcriptional activation
functions that can activate transcription when fused to a heterologous DNA-binding
domain.
The main determinants for transactivation map to NTD of both the androgen
(AR) and glucocorticoid (GR) receptors and while generally there is little sequence
conservation between the different NTD, in contrast to the DBD and LBD, short
regions of similarity have been observed for the AR and GR.[17]
The NTD is potentially involved in multiple protein-protein interactions and
the length of this domain has a positive correlation with the activity of AF-1
for different members of the nuclear receptor superfamily.[18]
C REGION (DNA BINDING DOMAIN, DBD)
The DBD consists of a highly conserved residue core located between the N-terminal
domain and the C-terminal ligand-binding domain. The DNA binding domain lies
toward the center of the molecule. The amino acid sequence of this domain is
similar among different steroid receptors (56–79% identity). The 3D structure
of the DBD has been resolved for a number of nuclear receptors. Nuclear magnetic
resonance and crystallographic studies for different NR DBD in their DNA uncomplexed
and complexed forms with the GR and ER homodimers on their cognate DNA sequence
were the first 3D crystal structure reported.[19]
D REGION (HINGE REGION)
The D region, which is a poorly conserved domain, serves as a hinge between
the DBD and the LBD, allowing rotation of the DBD. The hinge region allows the
DBD and LBD to adopt different conformations without creating a steric hindrance.
This domain also harbors a nuclear localization signal (NLS) or at least some
elements of a functional nuclear localization signal.
E REGION (LIGAND BINDING DOMAIN, LBD)
The largest domain is the moderately conserved ligand-binding domain (LBD,
E region). The hallmark of a nuclear receptor is its ligand-binding domain (LBD).
This domain is highly structured, and encodes a wealth of distinct functions
most of which operate in a ligand-dependent manner. The highly conserved region
of the nuclear receptor proteins lies near the carboxyl terminus. The AR C-terminal
ligand-binding domain contains about 290 amino acids and represents about 30%
of the receptor. The ligand-binding domains of AR from humans, rats, and mice
are identical, and sequence homology with other steroid receptors ranges between
15% and 54%.
Members of the NR superfamily (GR, MR, PR, AR, and ER) interact with heat-shock
proteins via their LBD.[20] The
LBD is a region where chaperone proteins, such as Hsp90, bind NR when they are
present in the cytoplasm. Upon ligand binding, exposure of a NLS in the LBD
induces the nuclear translocation of the NR. NR-LBD together with NR-DBD also
consists of a sequence involved in NR dimerization with the NR partner.[21]
After dissociation of chaperones, the liganded NR–complexes can bind to chromatin
organized DNA sequences near target genes, termed hormone response elements
(HRE). The HRE-recruited hormone-receptor-complexes are then able to initiate
chromatin remodeling and to relay activating or repressing signals to the target
genes transcription machinery.
Nuclear receptor pharmacology has led the way in the appreciation that ligands
may exert very diverse pharmacology, based on their individual chemical structure
and the allosteric changes induced in the receptor/accessory protein complex.[22]
Binding of agonistic or antagonistic ligands leads to different allosteric changes
of NR making them competent to exert positive or negative effects on the expression
of target genes by different mechanisms. In the upcoming articles, discussion
of these interactions is with a focus on the androgen receptor. The past decade
has seen a revolution in the study and investigation of androgen receptor ligands.
This brief introduction of the nuclear receptor serves as background for a more
complete understanding of new and revolutionary drugs currently under study
for clinical use.
References
[1]
The recommended usage of terms in the field of nuclear receptors is the
following: Isoforms are a product of the same gene produced by alternative
splicing, alternative promoter usage, or alternative translational initiation
and does not consider post-translational modifications. Subtypes are products
of related (paralogous) genes and use of this term is preferable to isotype.
Unliganded receptor is the preferable term to aporeceptor. Transactivation
is activation of transcription by the binding of a transcription factor
(and coregulators) to a DNA regulatory sequence. Coregulators are macromolecules
that associate with NR to modulate their transcriptional activity and are
divisible into coregulators that promote positive activity (coactivators)
and those that promote negative activity (corepressors). Ligands for NR
are compounds that bind reversibly to NR into the C-terminal LBP. For nuclear
receptors, the concept of agonist and antagonist is response and gene specific.
Agonists are ligands that induce an active conformation of the receptor.
Antagonists are ligands that produce a conformation and an action of the
receptor distinct from that produced by an agonist. Selective agonist and
antagonist are ligands with an affinity difference between their primary
target and other receptors. Partial agonists are agonists that in a given
tissue, under specific conditions, cannot elicit as optimal an effect as
can another agonist acting through the same receptors in the same tissue.
Inverse agonists are ligands that can promote corepressor recruitment. Potency
is an expression of the activity of a drug, in terms of the concentration
or amount needed to produce a defined effect—an imprecise term that should
always be further defined; drug potency depends on both receptor (affinity,
efficacy) and tissue (receptor numbers, drug accessibility) parameters;
term is sometimes incorrectly used to refer to the maximum effect attainable.
[2]
Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily.
Essays Biochem 2004;40:11–26.
[3]
Chambon P. The nuclear receptor superfamily: a personal retrospect on the
first two decades. Mol Endocrinol 2005;19:1418-28. Evans RM. The nuclear
receptor superfamily: a Rosetta stone for physiology. Mol Endocrinol 2005;19:1429-38.
Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the
nuclear receptor superfamily. Nat Rev Drug Discov 2004;3:950-64.
[4]
Nuclear Receptors Nomenclature Committee. A unified nomenclature system
for the nuclear receptor superfamily. Cell 1999;97:1 -3.
[5]
The Nuclear Receptor Group. Available at:
http://www.cbs.cnrs.fr/MAJ/SP/W.Bourguet/Website/research_focus.html
[6]
Escriva H, Safi R, Hanni C, et al. Ligand binding was acquired during evolution
of nuclear receptors. Proc Natl Acad Sci USA 1997;94:6803-8.
Owen GI, Zelent A. Origins and evolutionary diversification of the nuclear
receptor superfamily. Cell Mol Life Sci 2000;57:809-27.
[7]
Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature
of nuclear receptors. Pharmacol Rev 2006;58:685-704. Robinson-Rechavi M,
Escriva Garcia H, Laudet V. The nuclear receptor superfamily. J Cell Sci
2003;116:585-6.
[8]
In genetics, complementary DNA (cDNA) is DNA synthesized from a mature mRNA
template in a reaction catalyzed by the enzyme reverse transcriptase. The
central dogma of molecular biology outlines that in synthesizing proteins,
DNA transcribes into mRNA (transcription), and protein synthesis uses the
information in mRNA as a template (translation). Eukaryotic genes can contain
introns (intervening sequences), which are not coding sequences, and must
be spliced out of the RNA primary transcript before it becomes mRNA and
can be translated into protein. Complementary DNA is DNA spliced of the
introns (intervening sequences). Synthesis of cDNA is most often from mature
(fully spliced) mRNA using the enzyme reverse transcriptase. This enzyme
operates on a single strand of mRNA, generating its complementary DNA based
on the pairing of RNA base pairs (A, U, G and C) to their DNA complements
(T, A, C and G respectively).
[9]
Evans RM. The steroid and thyroid hormone receptor superfamily. Science
1988;240:889-95. Forman BM, Samuels HH. Interactions among a subfamily of
hormone receptors: the regulatory zipper model. Mol Endocrinol 1990;4:1293-301.
[10]
Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature
of nuclear receptors. Pharmacol Rev 2006;58:685-704. Nuclear Receptors Nomenclature
Committee. A unified nomenclature system for the nuclear receptor superfamily.
Cell 1999;97:1 -3.
[11]
Escriva H, Delaunay F, Laudet V. Ligand binding and nuclear receptor evolution.
Bioessays 2000;22:717-27. Nuclear Receptor Nomenclature Committee. A unified
nomenclature system for the nuclear receptor superfamily. Cell 1999;97:161-3.
Thornton JW, DeSalle R. A new method to localize and test the significance
of incongruence: detecting domain shuffling in the nuclear receptor superfamily.
Syst Biol 2000;49:183-201.
[12]
Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure:
implications for function. Annu Rev Physiol 2007;69:201-20. Germain P, Staels
B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear
receptors. Pharmacol Rev 2006;58:685-704.
[13]
Giguere V, Hollenberg SM, Rosenfeld MG, Evans RM. Functional domains of
the human glucocorticoid receptor. Cell 1986;46:645-52. Krust A, Green S,
Argos P, et al. The chicken oestrogen receptor sequence: homology with v-erbA
and the human oestrogen and glucocorticoid receptors. EMBO (Eur Mol Biol
Organ) J 1986;5:891-7.
[14]
Tata JR. Signaling through nuclear receptors. Nat Rev Mol Cell Biol 2002;3:702-10.
Robinson-Rechavi M, Garcia HE, Laudet V. The nuclear receptor superfamily.
J Cell Sci 2003;116:585-6. Bain DL, Heneghan AF, Connaghan-Jones KD, Miura
MT. Nuclear receptor structure: implications for function. Ann Rev Physiol
2006;69:9.1-9.2.
[15]
Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol
Rev 2001;81:1269–304. Nagpal S, Friant S, Nakshatri H, Chambon P. RARs and
RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2)
and heterodimerization in vivo. EMBO J 1993;12:2349–60.
[16]
Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol
Rev 2001;81:1269-304. Germain P, Staels B, Dacquet C, Spedding M, Laudet
V. Overview of nomenclature of nuclear receptors. Pharmacol Rev 2006;58:685-704.
[17]
Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation
domains: induction of active conformations. Biochem J 2005;391:449-64.
[18]
Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation
domains: induction of active conformations. Biochem J 2005;391:449-64. He
B, Bai S, Hnat AT, et al. An androgen receptor NH2-terminal conserved motif
interacts with the COOH terminus of the Hsp70-interacting protein (CHIP).
J Biol Chem 2004;279:30643-53.
[19]
Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic
analysis of the interaction of the glucocorticoid receptor with DNA. Nature
(Lond) 1991;352:497-505. Schwabe JW, Chapman L, Finch JT, Rhodes D. The
crystal structure of the estrogen receptor DNA-binding domain bound to DNA:
how receptors discriminate between their response elements. Cell 1993;75:567-78.
[20]
Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein
and immunophilin chaperones. Endocr Rev 197;18:306-60.
[21]
Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol
Rev 2001;81:1269-304. Kumar S, Saradhi M, Chaturvedi NK, Tyagi RK. Intracellular
localization and nucleocytoplasmic trafficking of steroid receptors: an
overview. Mol Cell Endocrinol 2006;246:147-56.
[22]
Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature
of nuclear receptors. Pharmacol Rev 2006;58:685-704.